A Python CLI tool for finding unused CIDR blocks in AWS VPCs
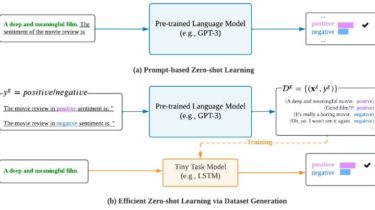
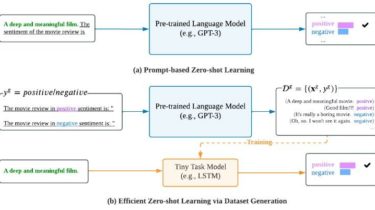
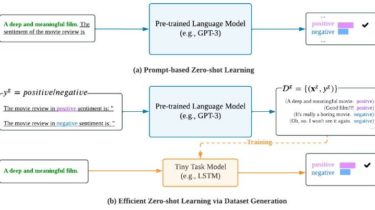
Overview aws-cidr-finder is a Python CLI tool which finds unused CIDR blocks (IPv4 only currently) in yourAWS VPCs and outputs them to STDOUT. It is very simple, but can be quite useful for users who managemany subnets across one or more VPCs. Use aws-cidr-finder -h to see command options. An Example It is easiest to see the value of this tool through an example. Pretend that we have the followingVPC setup in AWS: A VPC whose CIDR is 172.31.0.0/16, with […]
Read more