DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific Biosciences Circular Consensus Sequencing data
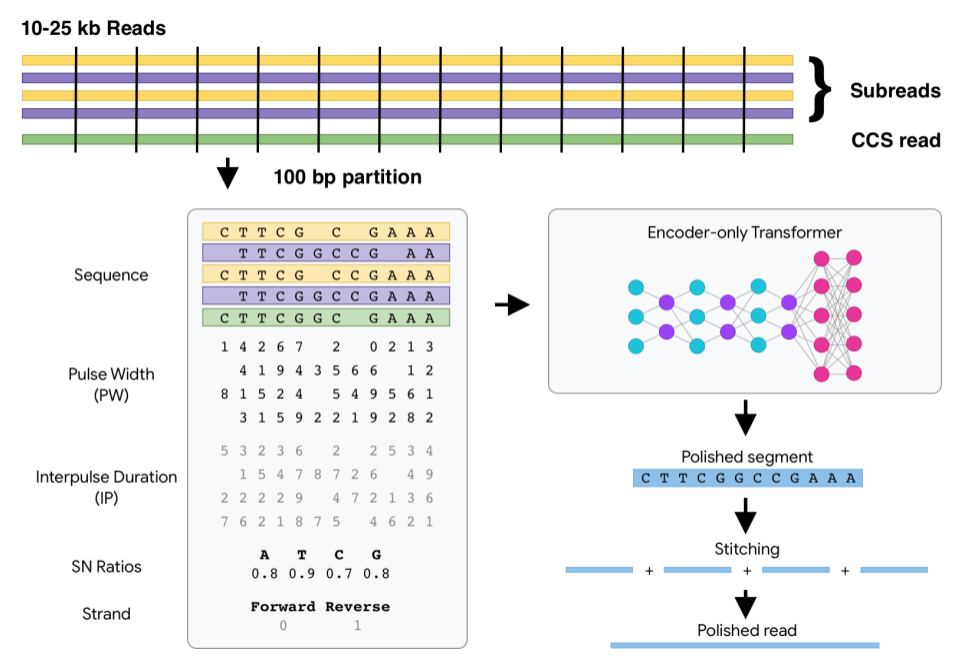
DeepConsensus uses gap-aware sequence transformers to correct errors in Pacific
Biosciences (PacBio) Circular Consensus Sequencing (CCS) data.
Installation
From pip package
pip install deepconsensus==0.1.0
You can ignore errors regarding google-nucleus installation, such as ERROR: Failed building wheel for google-nucleus.
From source
git clone https://github.com/google/deepconsensus.git
cd deepconsensus
source install.sh
(Optional) After source install.sh, if you want to run all unit tests, you can
do:
./run_all_tests.sh
Usage
See the quick start.
Where does DeepConsensus fit into my pipeline?
After a PacBio sequencing run, DeepConsensus is meant to be run on the CCS reads
and subreads to create new corrected reads in FASTQ format that can take the
place of the CCS reads for downstream analyses.
See the quick start for an example of inputs and outputs.
NOTE: This initial release