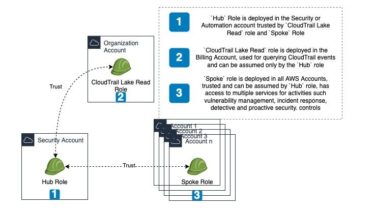
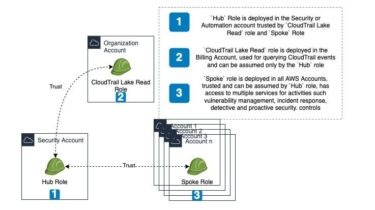
Gapmm2: gapped alignment using minimap2 (align transcripts to genome)
This tool is a wrapper for minimap2 to run spliced/gapped alignment, ie aligning transcripts to a genome. You are probably saying, yes minimap2 runs this with -x splice –cs option (you are correct). However, there are instances where the terminal exons from stock minimap2 alignments are missing. This tool detects those alignments that have unaligned terminal eons and uses edlib to find the terminal exon positions. The tool then updates the PAF output file with the updated information. Rationale We […]
Read more