A rotation-equivariant GNN for learning from biomolecular structure
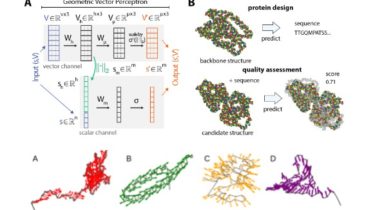
Geometric Vector Perceptron
Implementation of equivariant GVP-GNNs as described in Learning from Protein Structure with Geometric Vector Perceptrons by B Jing, S Eismann, P Suriana, RJL Townshend, and RO Dror.
This repository serves two purposes. If you would like to use the GVP architecture for structural biology tasks, we provide building blocks for models and data pipelines. If you are specifically interested in protein design as described in the paper, we provide scripts for training and testing models.
Note: This repository is an implementation in PyTorch Geometric emphasizing usability and flexibility. The original code for the paper, in TensorFlow, can be found here. We thank Pratham Soni for his contributions to the implementation in PyTorch.
Requirements
- UNIX environment
- python==3.6.13
- torch==1.8.1
- torch_geometric==1.7.0
- torch_scatter==2.0.6
- torch_cluster==1.5.9
- tqdm==4.38.0
- numpy==1.19.4
- sklearn==0.24.1
While